Infinite DMRG algorithm for the Heisenberg chain¶
The goal of this exercise is to implement the infinite version of the DMRG algorithm for the antiferromagnetic Heisenberg chain of spins one-half.
The Hamiltonian that we will build is the antiferromagnetic Heisenberg model:
![H=\sum_{i}\vec{S}_{i}\cdot\vec{S}_{i+1}=
\sum_{i}\left[S^{z}_{i}S^{z}_{i+1}+
\frac{1}{2}\left(S^{\dagger}_{i}S^{-}_{i+1}+
S^{-}_{i}S^{\dagger}_{i+1}\right)\right]](_images/math/19ccae83bf826a74d39cf31c7d02800c5939e603.png)
where 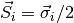 are the spin operators,
are the spin operators,
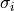 are the Pauli matrices, and
are the Pauli matrices, and
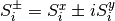 .
.
Exercise¶
Calculate the ground state energy of the antiferromagnetic Heisenberg
model for a chain of spins 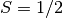 using the infinite version of
the DMRG algorithm. You should be able to pass the number of sites of the
chain and the number of states kept during the DMRG truncation as
parameters. The output should be the energy per site, entanglement entropy
and, truncation error at each step of the algorithm. You could:
using the infinite version of
the DMRG algorithm. You should be able to pass the number of sites of the
chain and the number of states kept during the DMRG truncation as
parameters. The output should be the energy per site, entanglement entropy
and, truncation error at each step of the algorithm. You could:
make a plot of the energy per site versus system size to see how converges. The relative error in energy is given by the truncation error.
for a given system size (say
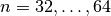 ) calculate the
energy per site for different number of states kept, make a plot, and
extrapolate the values of the energy to zero truncation error with a
linear fit. Compare this with the exact value for a finite size given by
the Bethe Anstatz.
) calculate the
energy per site for different number of states kept, make a plot, and
extrapolate the values of the energy to zero truncation error with a
linear fit. Compare this with the exact value for a finite size given by
the Bethe Anstatz.L E/J 16 -6.9117371455749 24 -10.4537857604096 32 -13.9973156182243 48 -21.0859563143863 64 -28.1754248597421
Solution¶
You can use the System class to create the four spin chain, and set the system Hamiltonian to the antiferromagnetic Heisenberg model, as we did in the previous exercise. Then you calculate ground state as before. From there you need to implement the DMRG transformation. We did most of the required steps (calculating, diagonalizing, and truncating the reduced density matrix) in a previous exercise.
The new thing is updating the operators using the transformation matrix.
You need to specify two things before doing that. First which are going to
be the operators to be transformed. These are the ones that you need in
the next step of the DMRG algorithm. To build the Heisenberg Hamiltonian
using the block-site-site-block construction, you need your block to have
a truncated version of the current single site spin operators
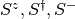 . Additionally you need to update the
block hamiltonian, which belongs to the current block and contains the
part of the Hamiltonian involving only degrees of freedom within the
block. This function sets the operators to be updated:
. Additionally you need to update the
block hamiltonian, which belongs to the current block and contains the
part of the Hamiltonian involving only degrees of freedom within the
block. This function sets the operators to be updated:
def set_operators_to_update_to_AF_Heisenberg(system):
"""Sets the operators to update to be what you need to AF Heisenberg.
Parameters
----------
system : a System.
The System you want to set the Hamiltonian for.
Notes
-----
The block Hamiltonian, althought needs to be updated, is treated
separately by the very functions in the `System` class.
"""
system.add_to_operators_to_update('s_z', site_op='s_z')
system.add_to_operators_to_update('s_p', site_op='s_p')
system.add_to_operators_to_update('s_m', site_op='s_m')
The block hamiltonian is built by including the previous block hamiltonian, if any, and the terms coming from the interactions between the block and the single site that is being included in the new block:
def set_block_hamiltonian_to_AF_Heisenberg(system):
"""Sets the block Hamiltonian to be what you need for AF Heisenberg.
Builds a matrix with the proper dimensions full of zeros. Then adds
the terms in the Hamiltonian that contain degrees of freedom belonging
only to one block. Finally adds the matrix to the block as the
operator labelled 'bh'.
Parameters
----------
system : a System.
The System you want to set the Hamiltonian for.
"""
tmp_matrix_size = None
if system.growing_side == 'left':
tmp_matrix_size = system.get_left_dim()
else:
tmp_matrix_size = system.get_right_dim()
tmp_matrix_for_bh = np.zeros((tmp_matrix_size, tmp_matrix_size))
if 'bh' in system.growing_block.operators.keys():
system.add_to_block_hamiltonian(tmp_matrix_for_bh, 'bh', 'id')
system.add_to_block_hamiltonian(tmp_matrix_for_bh, 's_z', 's_z')
system.add_to_block_hamiltonian(tmp_matrix_for_bh, 's_p', 's_m', .5)
system.add_to_block_hamiltonian(tmp_matrix_for_bh, 's_m', 's_p', .5)
system.operators_to_add_to_block['bh'] = tmp_matrix_for_bh
Now we can put together the function to grow a block by one site:
def grow_block_by_one_site(growing_block, ground_state_wf, system,
number_of_states_kept):
"""Grows one side of the system by one site.
Calculates the truncation matrix by calculating the reduced density
matrix for `ground_state_wf` by tracing out the degrees of freedom of
the shrinking side. Then updates the operators you need in the next
steps, effectively growing the size of the block by one site.
Parameters
----------
growing_block : a string.
The block which is growing. It must be 'left' or 'right'.
ground_state_wf : a Wavefunction.
The ground state wavefunction of your system.
system : a System object.
The system you want to do the calculation on. This function
assumes that you have set the Hamiltonian to something.
number_of_states_kept : an int.
The number of states you want to keep in each block after the
truncation. If the `number_of_states_kept` is smaller than the
dimension of the current Hilbert space block, all states are kept.
Returns
-------
entropy : a double.
The Von Neumann entropy for the cut that splits the chain into two
equal halves.
truncation_error : a double.
The truncation error, i.e. the sum of the discarded eigenvalues of
the reduced density matrix.
Raises
------
DMRGException
if `growing_side` is not 'left' or 'right'.
"""
if growing_block not in ('left', 'right'):
raise DMRGException('Growing side must be left or right.')
system.set_growing_side(growing_block)
rho = ground_state_wf.build_reduced_density_matrix(growing_block)
evals, evecs = diagonalize(rho)
truncated_evals, truncation_matrix = truncate(evals, evecs,
number_of_states_kept)
entropy = calculate_entropy(truncated_evals)
truncation_error = calculate_truncation_error(truncated_evals)
set_block_hamiltonian_to_AF_Heisenberg(system)
set_operators_to_update_to_AF_Heisenberg(system)
system.update_all_operators(truncation_matrix)
return entropy, truncation_error
A step of the infinite DMRG algorithm consists in growing at the same time both the left and the right blocks by one site.
def infinite_dmrg_step(system, current_size, number_of_states_kept):
"""Performs one step of the infinite DMRG algorithm.
Calculates the ground state of a system with a given size, then
performs the DMRG transformation on the operators of *both* blocks,
therefore increasing by one site the number of sites encoded in the
Hilbert space of each blocks, and reset the blocks in the system to be
the new, enlarged, truncated ones.
In reality the second block is not updated but just copied over from
the first.
Parameters
----------
system : a System object.
The system you want to do the calculation on. This function
assumes that you have set the Hamiltonian to something.
current_size : an int.
The number of sites of the chain.
number_of_states_kept : an int.
The number of states you want to keep in each block after the
truncation. If the `number_of_states_kept` is smaller than the
dimension of the current Hilbert space block, all states are kept.
Returns
-------
energy_per_site : a double.
The energy per site for the `current_size`.
entropy : a double.
The Von Neumann entropy for the cut that splits the chain into two
equal halves.
truncation_error : a double.
The truncation error, i.e. the sum of the discarded eigenvalues of
the reduced density matrix.
Notes
-----
Normally you don't update both blocks. If the chain is symmetric, you
just can use the operators for the one of the sides to mirror the
operators in the other side, saving the half of the CPU time. In
practical DMRG calculations one uses the finite algorithm to
improve the result of the infinite algorithm, and one of the blocks
is kept one site long, and therefore not updated.
"""
set_hamiltonian_to_AF_Heisenberg(system)
ground_state_energy, ground_state_wf = system.calculate_ground_state()
entropy, truncation_error = grow_block_by_one_site('left', ground_state_wf,
system,
number_of_states_kept)
system.right_block = system.left_block
return ground_state_energy / current_size, entropy, truncation_error
The other thing remaining is to make a small change in the function to set up the AF Heisenberg Hamiltonian that we used in the previous exercise to include the block hamiltonians:
def set_hamiltonian_to_AF_Heisenberg(system):
"""Sets a system Hamiltonian to the AF Heisenberg Hamiltonian.
Does exactly this. If the system hamiltonian has some other terms on
it, there are not touched. So be sure to use this function only in
newly created `System` objects.
Parameters
----------
system : a System.
The System you want to set the Hamiltonain for.
"""
system.clear_hamiltonian()
if 'bh' in system.left_block.operators.keys():
system.add_to_hamiltonian(left_block_op='bh')
if 'bh' in system.right_block.operators.keys():
system.add_to_hamiltonian(right_block_op='bh')
system.add_to_hamiltonian('id', 'id', 's_z', 's_z')
system.add_to_hamiltonian('id', 'id', 's_p', 's_m', .5)
system.add_to_hamiltonian('id', 'id', 's_m', 's_p', .5)
system.add_to_hamiltonian('id', 's_z', 's_z', 'id')
system.add_to_hamiltonian('id', 's_p', 's_m', 'id', .5)
system.add_to_hamiltonian('id', 's_m', 's_p', 'id', .5)
system.add_to_hamiltonian('s_z', 's_z', 'id', 'id')
system.add_to_hamiltonian('s_p', 's_m', 'id', 'id', .5)
system.add_to_hamiltonian('s_m', 's_p', 'id', 'id', .5)
Now you just make a loop that repeats the DMRG step until you reach the desired system size. Adding some code to pass the arguments from the command line and to save the results to a file, you get something like this:
def main(args):
#
# create a system object with spin one-half sites and blocks.
#
spin_one_half_site = SpinOneHalfSite()
system = System(spin_one_half_site)
#
# read command-line arguments and initialize some stuff
#
number_of_sites = int(args['-n'])
number_of_states_kept= int(args['-m'])
sizes = []
energies = []
entropies = []
truncation_errors = []
#
# infinite DMRG algorithm
#
number_of_sites = 2 * (number_of_sites / 2) # make it even
for current_size in range(4, number_of_sites + 1, 2):
#block_size = current_size / 2 - 1
energy, entropy, truncation_error = (
infinite_dmrg_step(system, current_size, number_of_states_kept) )
sizes.append(current_size)
energies.append(energy)
entropies.append(entropy)
truncation_errors.append(truncation_error)
#
# save results
#
output_file = os.path.join(os.path.abspath(args['--dir']), args['--output'])
f = open(output_file, 'w')
zipped = zip (sizes, energies, entropies, truncation_errors)
f.write('\n'.join('%s %s %s %s' % x for x in zipped))
f.close()
print 'Results stored in ' + output_file
See a full implementation of the above code. To learn how to run this code you
can use:
$ ./tutorial/solutions/infinite_heisenberg.py --help
Implements the infinite version of the DMRG algorithm for the S=1/2 AF
Heisenberg.
Calculates the ground state energy and wavefunction for the
antiferromagnetic Heisenberg model for a chain of spin one-half. The
calculation of the ground state is done using the infinite version of the
DMRG algorithm.
Usage:
infinite_heisenberg.py (-m=<states> -n=<sites>) [--dir=DIR -o=FILE]
infinite_heisenberg.py -h | --help
Options:
-h --help Shows this screen.
-n <sites> Number of sites of the chain.
-m <states> Number of states kept.
-o --output=FILE Ouput file [default: infinite_heisenberg.dat]
--dir=DIR Ouput directory [default: ./]